Mountain Birdwatch Overview (2010 to present)
Mountain Birdwatch is focused on the breeding bird community, and its primary nest predator (Red Squirrel) within the high-elevation spruce-fir forests of the northeastern United States (Eastern New York and Northern New England). To establish long-term sampling stations back in 2010, we used a model of the Bicknell’s Thrush breeding habitat and range (based on Lambert et al. 2005; for an interactive map of that range, click here) as a proxy for the extent of spruce-fir boreal habitat in the northeastern U.S. Incredibly, there isn’t simply a GIS layer of the complete spruce-fir extent within our region. As such, most of the sampling stations occur solidly in the spruce-fir zone, but some stations occur in the hardwood-boreal transition or at treeline (which is incredibly valuable for the monitoring of some of our species). We used generalized random tessellation stratified sampling (GRTS) to select blocks of the spruce-fir zone to establish survey routes and ~750 sampling stations. Sampling stations occur between 573 and 1500 meters (mean = 1009 meters) in elevation, but 95% of the sampling stations occur between 759 and 1333 meters. Each Mountain Birdwatch route consists of 3-6 sampling stations located on established hiking trails (or occasionally retired logging roads) for safety purposes, as these habitats are incredibly remote and rugged. The number of sampling stations on a route was determined by the size of the spruce-fir zone on that mountain and the extent of the hiking trail network. Over time, a few routes and individual sampling stations have been permanently retired (e.g., due to safety concerns and trail closures), and new sampling stations have been added to existing routes (e.g., along sections of newly-built trails). As of 2022, a total of 791 Mountain Birdwatch sampling stations have been active at some point since 2010.
Each year, community scientists (birders who like to hike, or hikers who like to bird) adopt one or more routes to survey on any day in June with fair weather. While some of our community scientists are professional biologists and birding guides, most are simply naturalists from all walks of life. Mountain Birdwatch intentionally features a short list of species to survey, and extensive training materials for participants, so just about any birder can contribute with practice and training. Most observers actually record complete checklists (of all bird species) for Mountain Birdwatch, and those data are entered into our database, but we only analyze (at this point) observations of the 10 bird and one mammal species that all observers are only required to count. Most observers survey the same route year after year and become intimately familiar with ‘their’ route. We encourage observers to bring along a companion for safety purposes, but that companion does not assist the observer in detecting or counting birds in any capacity. Due to the remote sampling locations and early morning survey start time, most observers backcountry camp overnight prior to the survey near their first sampling station.
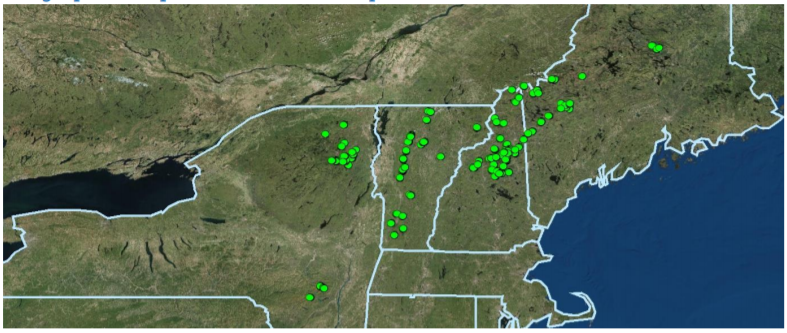
All ~750 Mountain Birdwatch sampling stations.
Species and Survey Methods
Only a few dozen bird species regularly breed in the spruce-fir zone in our region. From this group, we selected ten bird species for targeted monitoring based on level of conservation concern, degree of habitat specialization, probability of upslope range expansion, and range restriction. We also monitor the abundance of red squirrels to understand how this common nest predator’s cyclical population dynamics interact with those of our monitored bird species.
We designed the Mountain Birdwatch surveys to achieve three main goals, to:
- estimate on an annual basis the abundance of focal species within the mountains of our region,
- measure changes in the abundance of focal species over time, and
- relate trends in abundance to biotic and abiotic variables that may affect the focal species.
The Mountain Birdwatch protocol consists of four consecutive 5-minute independent counts at each sampling station, for a total sampling period of 20 minutes per station. Observers conduct repeated counts for all 11 focal species during each 5-minute period, noting whether individuals were detected within or beyond 50 meters of the station. Since 2019, observers can optionally record counts for any bird species (not just the focal species) and indicate if their 5-minute counts are complete checklists. This repeated measures sampling format is one of the greatest strengths of our sampling protocol. By conducting multiple back-to-back counts, we can statistically account for individual birds missed by observers during their counts. Surveys are conducted during the month of June, which is the collective period of greatest vocal activity for the focal species. Observers begin surveys 45 minutes before local sunrise and surveys are typically finished before 8 am. The vast majority of detections (~98%) are made by ear, as the forest habitat is typically quite dense. Note, observers formerly recorded whether detections were initially by ear or by sight. Inclement weather can greatly reduce an observer’s ability to detect birds in the field, so each survey is conducted during fair weather (wind speeds <19 kph, >0℃, and no rain).
Analyzing the Data
Community scientists enter their own survey data into a sophisticated online database (managed by the Forest Ecosystem Monitoring Cooperative) and mail in their original datasheets. Each summer, our interns proof these datasheets (and the online data entries) for transcription errors and unusual observations–contacting the observers when needed. The Mountain Birdwatch Chief Scientist, Jason Hill, is a quantitative ecologist who currently analyzes Mountain Birdwatch data using hierarchical binomial N-mixture models in a Bayesian framework. N-mixture models estimate local abundance after accounting for imperfect detection by observers, and can easily handle missing count data if a particular route isn’t surveyed in a particular year.
Each species is analyzed separately, using the full suite of the Mountain Birdwatch data from 2010 to the current year with two exceptions–Fox Sparrow and Boreal Chickadee models only include data from sampling stations located north of 43.5 degrees latitude (the approximate southern breeding boundary of both species in our region). For the annual State of the Birds Report, we run the same N-mixture model for each focal species using program R and JAGS. On the abundance side of the equation, the models contain annual and regional intercepts, a linear time trend, a random year effect, and univariate and quadratic forms of elevation and latitude as covariates. The detection side of the equation contains an annual intercept and univariate and quadratic forms of a covariate representing the point count start time converted to minutes before (negative values) or after (positive values) local sunrise time, wind speed, and ordinal date. All covariates are grand-mean centered and scaled by 1 SD.
Final inferences are drawn from the posteriors of a model with 3000 saved iterations (1000 iterations saved from three Markov chain Monte Carlo processes) after a thinning rate of 1 in 10 [see Link and Eaton for a reminder about the uselessness of heavy thinning rates].
In all models we specify a uniform prior on detection probability (min=0, max=1), weakly informative logistic priors centered at 0 with scale parameter 1 (~ dlogis(0,1)) for intercepts and covariate coefficients modeling local abundance and detection (Northrup and Gerber 2018), and a uniform (min=0,max=1) prior for the standard deviation of the random year effect. Each year we conduct a sensitivity analysis on the priors for intercepts and covariate coefficients by comparing the mean and standard errors of parameters produced via 1) packaged unmarked (i.e., maximum likelihood estimates), 2) normally distributed priors with mean zero and a variance of 2, 10, or 20, and 3), our weakly informative logistic priors. From each model, we retained 3000 total iterations from 3 Markov chain Monte Carlo chains (MCMC), thinned at a rate of 1:5, with a burn-in of 5000 iterations. For each species’ model results, we examine plots of the residuals and traceplots of the posteriors and verify that Gelman-Rubin statistics are <1.01 (Gelman and Rubin 1992).
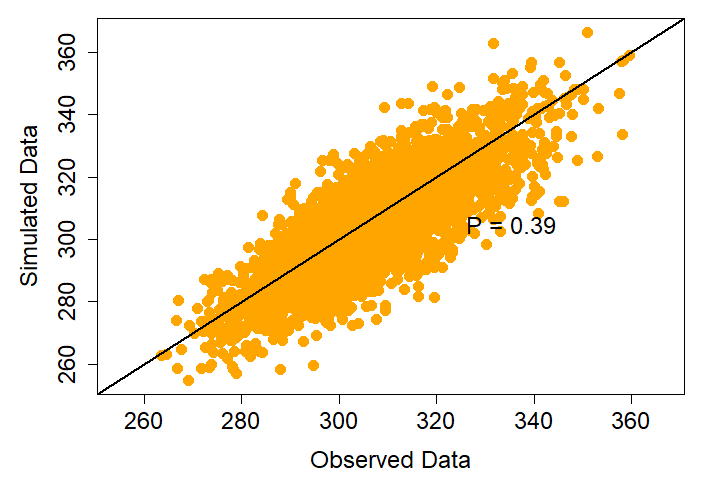
Posterior predictive check. All models are predictive…but are they good at it? From each species’ model, we generate and compare 100,000 simulated data sets to our observed data set to insure that our model makes accurate predictions. The ratio of the simulated data that over- and under-predict the observed data should be P = 0.5 (in a perfect world). Here, P = 0.39 for our Fox Sparrow model likely indicates a well-fitting model that makes reasonable predictions.


